How to access the TOFF database and download its content using R
A) Download the R script and launch R
The R script needed to access the database can be downloaded here. If you already installed R, you may now launch it and open the script.
B) Open the script via R
In R : Click on “File” in the top left corner of the software and then click “Open script” in the menu that appeared.

You should now see the script in a new window.
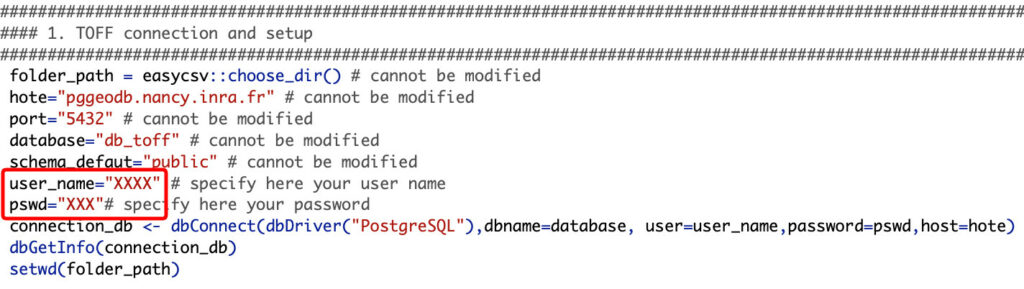
C) Login and password
The script is divided into several parts, clearly labeled in the code. In the part called “TOFF connection and setup” (part 1), you should see a field “user_name” and a field “pswd”. You may replace the placeholder between quotation with your login and your password, respectively in the “user_name” and “pswd” fields.
D) Running the script
To run the script, ensure that you have filled all the necessary fields (login, password) before doing so or the script will result in an error. Before running the script, you also need to ensure that your machine is connected to the internet.
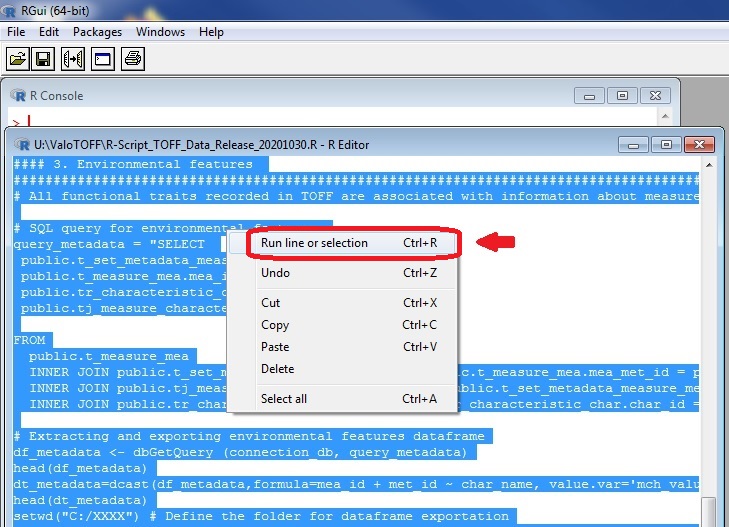
In R, from the R Editor window, select all the script and right click on it. Then, in the menu that appeared, click “Run line or selection”.
The first time you run the script, a window will appear asking you to specify which mirror you wish to use to download libraries. You may choose the location closest to your actual location. Another window will soon follow, asking you to specify where you want to save the files that will be downloaded.
E) Downloaded files
When the script is done, you will get six files at the location that you specified: “df_traits” in csv and xlsx, “df_metadata” in csv and xlsx and “df_merged_metadata_traits” in csv and xlsx. All functional traits recorded in TOFF (df_traits) are associated with information about measurements in an environmental context (df_metadata). The link between “df_traits” and “df_metadata” can be established through the “mea_id” or “met_id” columns or simply by looking at the merged file “df_merged_metadata_traits”. More information on the content of the database can be found in the “information” section of the website, or in the published paper (DOI: 10.1038/s41597-019-0307-z).